Le jeudi 16 mai 2024, Benjamin LAMBERT soutiendra sa thèse intitulée "Quantification de l'incertitude des réseaux profonds pour la segmentation d'images médicales".
Cette thèse a été dirigée par Michel DOJAT de l'équipe "Neuroimagerie Fonctionnelle et Perfusion Cérébrale" du GIN, Florence FORBES de l'INRIA et Senan DOYLE de Pixyl.
Composition du jury :
- Dr. Maria Zuluaga, Assistant Professor, Eurecom - Rapporteure
- Dr. Ninon Burgos, CNRS researcher, Institut du Cerveau - Rapporteure
- Pr. Pierrick Coupé, Professor, CNRS Délégation Aquitaine - Examinateur
- Dr. Julien Mairal, Research Scientist, INRIA Grenoble - Examinateur
- Dr. Christian Baumgartner, Assistant Professor, University of Lucerne - Examinateur
- Pr. Olivier François, Professor, Grenoble INP - Examinateur
Résumé :
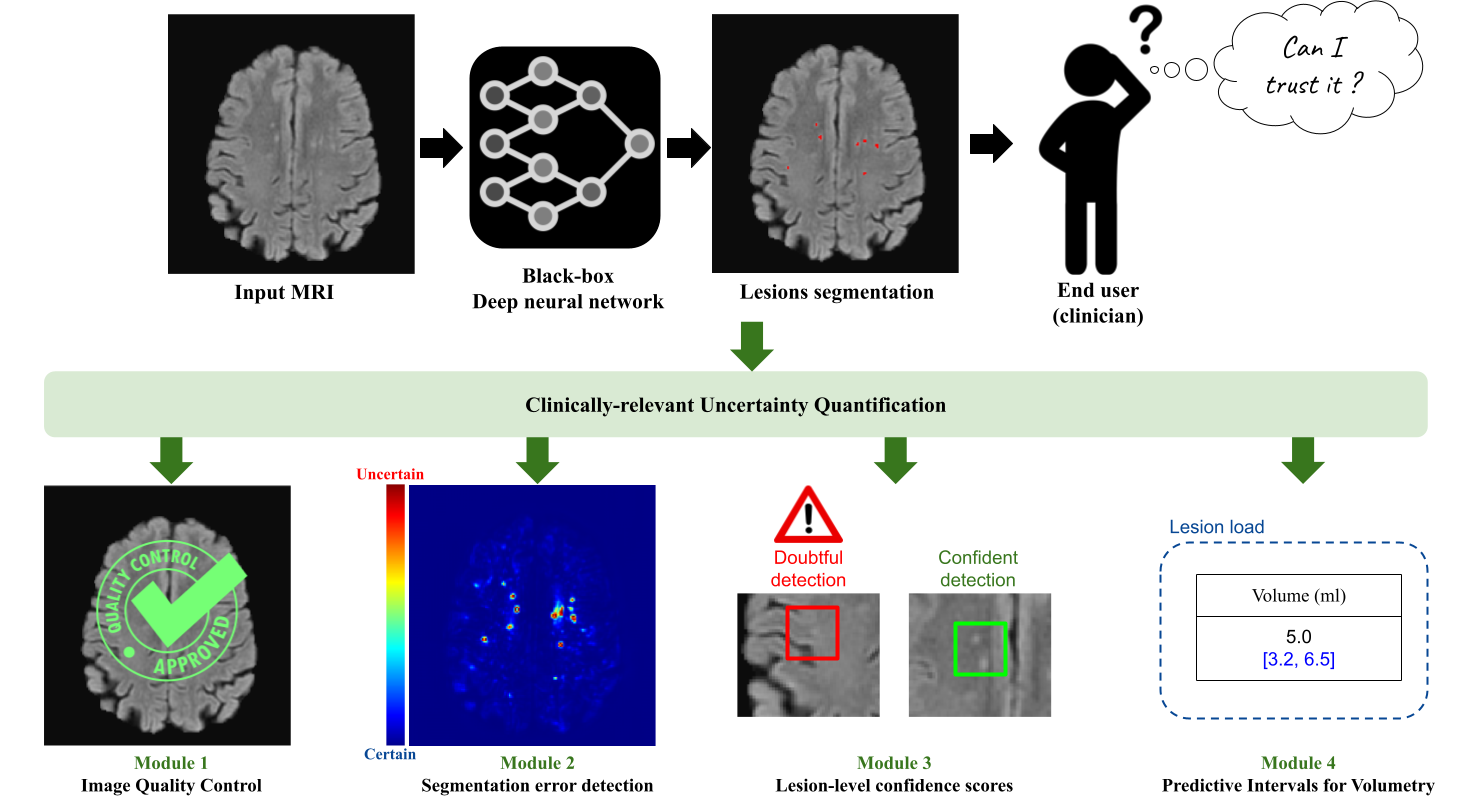
Quantifying the inherent uncertainty in an automated medical image analysis is crucial to guarantee the safe deployment of deep learning models. However, these models, often referred to as black boxes, are known to produce errors with high confidence, potentially leading to misinformed conclusions. The operative goal of Pixyl, GIN, and Inria for this Ph.D. is to develop flexible uncertainty quantification tools that would address the opacity of deep learning models. In medical image analysis, uncertainty estimates are useful at different levels: the voxel, the lesion, the subject (input image or output segmentation), and the estimated volumes.
At the voxel level, there are a multitude of solutions proposed in the literature. Through three different brain lesion segmentation tasks, we show the adequacy of the Deep Ensemble framework to detect incorrectly classified voxels. The resulting uncertainty map can be overlayed on the input image to identify the ambiguous regions. Second, specifically tailored for diseases involving the detection of multiple lesions such as Multiple Sclerosis, we propose a lesion-level uncertainty module that associates each identified brain lesion with a confidence score. This allows to draw the clinician's attention to these uncertain lesion instances, which may correspond to false positive detections. Then, as deep learning models lack robustness when the test image is not represented in the training dataset, we build two different subject-level quality control tools. Non-conform inputs are detected using a compressed latent representation, allowing to efficiently compute its distance to the training distribution in a low-dimensional manifold. Yet, this approach focuses on the conformity of the input image and thus is unhelpful in detecting poor-quality segmentations. To alleviate this, a second quality check focusing on segmentation quality is implemented, allowing to enrich the informativeness of the case-level uncertainty quantification. Lastly, we leverage the conformal prediction framework to equip lesional volume estimations with robust predictive intervals. The proposed framework, TriadNet, computes the segmentation and associated predictive intervals in a second, thus being ideal for industrial software.
The tools developed during this PhD will allow to enhance medical image analysis software with useful uncertainty estimates, allowing increased trust in the automated results and enabling informed decision-making.